RCSB Protein Data Bank
The Worldwide Protein Data Bank (wwPDB) consists of organizations that act as deposition, data processing and distribution centers for PDB data. The founding members are RCSB PDB (USA), PDBe (Europe) and PDBj (Japan) 1. The BMRB (USA) group joined the wwPDB in 2006. The mission of the wwPDB is to maintain a single Protein Data Bank Archive of macromolecular structural data that is freely and publicly available to the global community.
Crystal structures of the Bracher Group:
The crystal structures done in collaboration with Prof. Stefan Knapp, Structural Genomics Consortium (SGC), Oxford (www.sgc.ox.ac.uk), with Prof. Oded Livnah, Jerusalem and with Prof. Bernhard Kuster
| In collaboration with Prof. Stefan Knapp: | ||
|
|
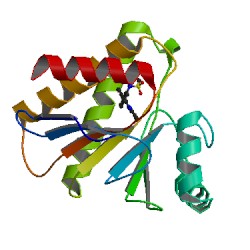
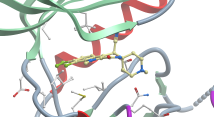
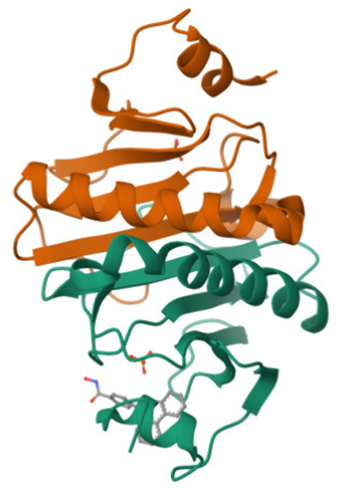
PARP14 Macrodomain 2 with inhibitor |
|
|
Authors: Uth, K., Schuller, M., Sieg, C., Wang, J., Krojer, T., Knapp, S., Riedels, K., Bracher, F., Edwards, A.M., Arrowsmith, C., Bountra, C., Elkins, J.M. DOI: 10.2210/pdb5O2D/pdb |
||
|
|
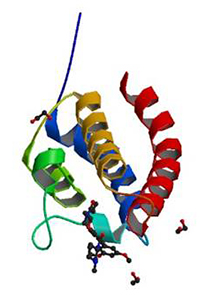
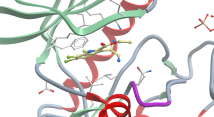
Crystal structure of the bromodomain of human CREBBP in complex with a benzoxazepine compound |
|
|
Authors: Tallant, C., Popp, T.A., Fedorov, O., Siejka, P., Picaud, S., von Delft, F., Arrowsmith, C.H., Edwards, A.M., Bountra, C., Bracher, F., Knapp, S., Structural Genomics Consortium DOI: 10.2210/pdb5j0d/pdb |
||
|
|
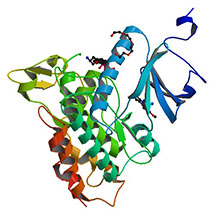
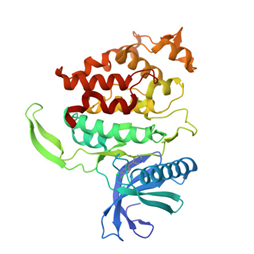
Crystal structure of DYRK1A with harmine-derivatized AnnH-75 inhibitor |
|
|
Authors: Chaikuad, A., Wurzlbauer, A., Nowak, R., von Delft, F., Arrowsmith, C.H., Edwards, A.M., Bountra, C., Bracher, F., Knapp, S. DOI: 10.2210/pdb4yu2/pdb |
||
|
|
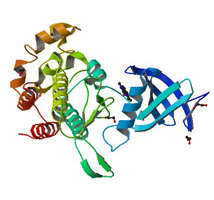
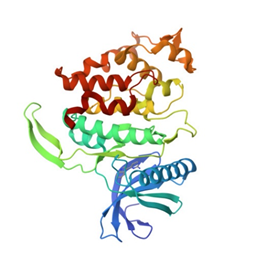
Crystal structure of Activin receptor type-IIA (ACVR2A) kinase domain in complex with a beta-carboline inhibitor |
|
|
Authors: Williams, E., Chaikuad, A., Canning, P., Kochan, G., Mahajan, P., Cooper, C.D.O., Beltrami, A., Krojer, T., Pohl, B., Bracher, F., Arrowsmith, C.H., Edwards, A.M., Bountra, C., Von Delfit, F., Bullock, A. DOI: 10.2210/pdb4asx/pdb |
||
|
|
Crystal structure of the first bromodomain of human BRD4 in complex with a benzo-triazepine ligand (BzT-7) |
|
|
Authors: Filippakopoulos, P., Picaud, S., Felletar, I., Fedorov, O., von Delft, F., Arrowsmith, C.H., Edwards, A.M., Weigelt, J., Bountra, C., Bracher, F., Knapp, S. DOI: 10.2210/pdb3u5l/pdb |
||
|
|
Crystal structure of the Human Clk3 in Complex with V25 (KH-CB19T) |
|
|
Authors: Muniz, J.R.C., Fedorov, O., King, O., Filippakopoulos, P., Bullock, A., Phillips, C., Heightman, T., Ugochukwu, E., Von Delft, F., Arrowsmith, C.H., Bracher, F., Huber, K., Edwards, A.M., Weigelt, J., Bountra, C., Knapp, S.
|
||
|
|
Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a beta carboline ligand II (KH-CARB13) |
|
|
Authors: Filippakopoulos, P., Bullock, A., Fedorov, O., Huber, K., Bracher, F., Pike, A.C.W., von Delft, F., Arrowsmith, C.H., Edwards, A.M., Bountra, C., Knapp, S.
|
||
|
|
Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a beta carboline ligand (KH-CARB10) |
|
|
Authors: Filippakopoulos, P., Bullock, A., Fedorov, O., Huber, K., Bracher, F., Pike, A.C.W., Roos, A., von Delft, F., Arrowsmith, C.H., Edwards, A.M., Bountra, C., Knapp, S.
|
||
|
|
Crystal structure of Human Death Associated Protein Kinase 3 (DAPK3) in Complex with a Beta-Carboline Ligand (KH-CARB10) |
|
|
Authors: Filippakopoulos, P., Rellos, P., Eswaran, J., Fedorov, O., Berridge, G., Niesen, F., Bracher, F., Huber, K., Pike, A.C.W., Roos, A., von Delft, F., Arrowsmith, C.H., Edwards, A.M., Weigelt, J., Knapp, S.
|
||
|
|
Crystal structure of Di-Phosphorylated Human Clk1 in Complex with a Novel Substituted Indole Inhibitor (KH-CB19T) |
|
|
Authors: Pike, A.C.W., Bullock, A., Fedorov, O., Pilka, E.S., Ugochukwu, E., Von Delft, F., Edwards, A., Arrowsmith, C.H., Weigelt, J., Sundstrom, M., Huber, K., Bracher, F., Knapp, S.
|
||
|
|
||
| In collaboration with Prof. Oded Livnah: | ||
|
|
Crystal structure of di-phosphorylated human CLK1 in complex with 5-(6,7-dichloro-1H-indol-3-yl)pyrimidin-4-amine |
|
|
Authors: Livnah, O., Domovich, Y., Bracher, F., Aigner, C.
|
||
|
|
Crystal structure of di-phosphorylated human CLK1 in complex with 4-(1H-indol-3-yl)pyrimidin-2-amine |
|
|
Authors: Livnah, O., Domovich, Y., Bracher, F., Aigner, C.
|
||
|
|
Non-phosphorylated human CLK1 in complex with an indole inhibitor to 1.65 Ang |
|
|
Authors: Livnah, O., Domovich, Y.
|
||
|
|
Crystal structure of di-phosphorylated human CLK1 in complex with 4-(6,7-dichloro-1H-indol-3-yl)pyrimidin-2-amine |
|
|
Authors: Livnah, O., Domovich, Y.
|
||
|
|
Crystal structure of di-phosphorylated human CLK1 in complex with 5-(1-methyl-1H-indol-3-yl)pyrimidin-4-amine |
|
|
Authors: Livnah, O., Domovich, Y.
|
||
|
|
Crystal structure of di-phosphorylated human CLK1 in complex with 4-(1-methyl-1H-indol-3-yl)pyrimidin-2-amine |
|
|
Authors: Livnah, O., Domovich, Y.
|
||
| In collaboration with Prof. Bernhard Kuster: | ||
|
|
Crystal structure of human Histidine Triad Nucleotide-Binding Protein 1 in complex with KV24 |
|
|
Authors: Dolot, R.M., Lechner, S., Sethiya, J.P., Wagner, C.R., Bracher, F., Kuster, B.
|
||
|
|
Crystal structure of human Histidine Triad Nucleotide-Binding Protein 1 in complex with KV30 |
|
|
Authors: Dolot, R.M., Lechner, S., Sethiya, J.P., Wagner, C.R., Bracher, F., Kuster, B.
|
||